Thus we conclude that Primer-BLAST is able to detect potential unintended targets that are missed by QuantPrime or PRIMERGENS during the specificity screening process. A different DNA template was used for the experiment presented in Figure 3b. Genomic DNA from a Mycobacteriophage was used to amplify a conserved 566 bp DNA segment. Like the previous experiment, the optimal Mg2+ concentration had to be determined. As shown in Figure 3b, amplification of the desired PCR product requires at least 2.0 mM Mg2+ (lane 5).
Steps
During the amplification process, through denaturation, the secondary structures will also be opened. It is important to check that all secondary structures have a Tm (°C) less than the qPCR annealing temperature (normally 55-60°C) to ensure effective amplification 68. Therefore, we suggest that in the primer design for SARS-CoV-2, it is more effective to design the primer targeting position on the stem, at least to make sure that the 3 ' terminal bases are on the stem (as shown in Figure Figure3).3). However, further experiments are needed to explore and verify this.
STEPS INVOLVED IN PERFORMING REAL-TIME PCR
Ideally, a primer pair should only amplify the intended target, but not any unintended targets. PCR is a commonly used method to amplify DNA of interest in many fields such as biomedical research, diagnostic testing and forensic testing. While the outcome of PCR can be influenced by many other conditions such as the template DNA preparation and reaction conditions, designing a good pair of primers is a critical factor. A general requirement is that primers should have similar melting temperatures (Tm) and a balanced G/C content, but should avoid self-complementarity and hair-pin structure. For example, to avoid unwanted amplification of genomic DNA in reverse transcription PCR (RT-PCR), it is recommended that a primer pair span an intron, or that one of the primers be located at an exon-exon junction. Another concern is the possible impact of SNPs in the primer regions.
Search
FBPP: software to design PCR primers and probes for nucleic acid base detection of foodborne pathogens Scientific ... - Nature.com
FBPP: software to design PCR primers and probes for nucleic acid base detection of foodborne pathogens Scientific ....
Posted: Fri, 12 Jan 2024 08:00:00 GMT [source]
This review also discusses the need for accurate diagnosis and timely treatment of the new coronavirus pneumonia. Users can specify the number of mismatches that a primer pair must have to unintended targets as well as a 3’ end region where these mismatches must be present. In addition, users can specify the mismatch threshold above which any targets should be ignored (i.e., filtering out targets having too many mismatches to be a concern for non-specific amplification). Several database options are available for specificity checking with broad organism coverage.
For example, if you want the PCR product to be located between position 100 and position 1000 on the template, you can set forward primer "From" to 100 and reverse primer "To" to 1000 (but leave the forward primer "To" and reverse primer "From" empty). Note that the position range of forward primer may not overlap with that of reverse primer. In addition to sensitivity, specificity is an important indicator when considering primers and probes. Whether there is cross-reactivity is an important aspect of evaluating specificity.
This is essential to ensure that the designed primers do not amplify non-specific products 38. At present, gene sequences of the six known coronaviruses (HCoV-229E, HCoV-NL63, HCoV-HKU1, HCoV-OC43, MERS-CoV, and SARS-CoV) have been published, and primers and probes can be designed based on the available genomic information. When an unknown coronavirus suddenly emerges and causes a new type of infectious pneumonia, how should we respond? In such a scenario, designing effective primers with high specificity and sensitivity can have immense value for diagnosis, tracking, and tracing.
Primer design for quantitative real-time PCR for the emerging Coronavirus SARS-CoV-2
The primers, probes, and reagents used for detection could be another critical factor. Finally, RT-qPCR primers are designed to amplify the target regions of the SARS-CoV-2 genome. However, the novel coronaviruses are RNA viruses, and once mutations and recombination occur, RT-qPCR primers will not be able to effectively amplify the viral sequences.
Results and Discussion

This requires at least one primer (for a given primer pair) to have the specified number of mismatches to unintended targets. The larger the mismatches (especially those toward 3' end) are between primers and the unintended targets, the more specific the primer pair is to your template (i.e., it will be more difficult to anneal to unintended targets). However, specifying a larger mismatch value may make it more difficult to find such specific primers.
A number of public software tools have been developed to aid the primer design process. Notably, the widely used Primer3 program [7] designs primers based on a variety of parameters. Since it does not perform target analysis, users typically need to test the primer specificity using additional tools. However, this process is time-consuming and sometimes even impractical if the primers have too many database matches (as with a BLAST search, for example). This is another parameter that can be used to adjust primer specificity stringecy. If the total number of mismatches between target and at least one primer (for a given primer pair) is equal to or more than the specified number (regardless of the mismatch locations), then any such targets will be ignored for primer specificity check.
Therefore, it is necessary to know as much information as possible related to the RNA genome sequences. The first step involves obtaining the maximum amount of information from sequence databases; it is crucial to refer to the accession or individual transcript number of related sequences for assay design. 5.3 Binding of primer to the complementary strand and extension of the primer. Polymerization results in the synthesis of the upper strand (5’-3’). Partly the problem comes from the fact that that way we communicate DNA sequences. The other half of the problem is that we have to work with the template strand to design a primer for the other strand.
There are several existing programs that have addressed some aspects of this issue. Autoprime [10] designs primers spanning exon junctions or introns so that the primers only target mRNA. QuantPrime [11] is a specialized tool to design target-specific primers for detecting mRNA in real time PCR. Likewise, the PRIMEGENS Sequence Specific Primer Design tool [12] can also be used for specific primer design for a limited number of organisms. Other limitations in these tools include low target detection sensitivity, limited specificity stringency options, no or limited support for designing primers based on exon/intron boundary requirements and limited coverage of organisms in search databases. Primer-BLAST is a general purpose target-specific PCR primer design tool that offers a level of sensitivity and usability not found in other tools.
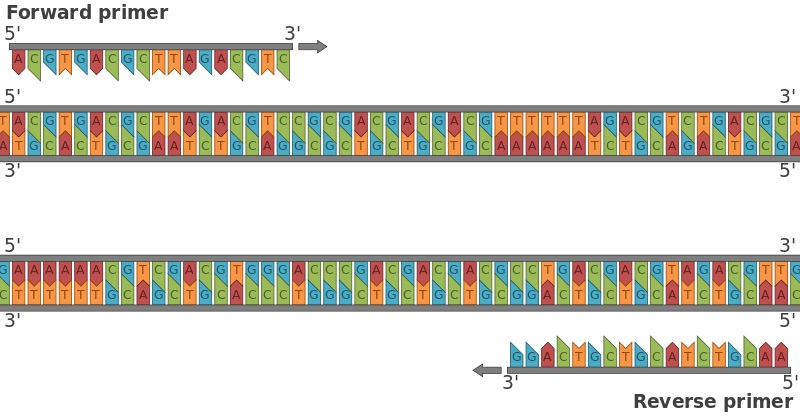
The 3′-OH ends of the primers face each other because they have annealed to opposite strands. The primers are then extended by DNA polymerase, creating two new double-stranded DNA molecules, each one containing a copy of the desired sequence or gene. Forward primers can also be called sense primers while the reverse primers are also known as the anti-sense primers. Primers generally bind to the opposite sides of the template DNA sequence or strands to be amplified during the PCR reaction. They are single-stranded (ss) DNA molecules or oligonucleotides, and they are synthesized by chemical processes.


No comments:
Post a Comment